3. Results
Power law hypothesis test on the out-degree distribution of a node
We analyzed the characteristics of the out-degree distribution of nodes to identify the features of the COVID-19 infection network in South Korea. The out-degrees were calculated considering the link direction [16].
First, the out-degree distribution of all nodes is presented in Figure 2. The pattern of the graph shows a model of distribution with heavy tails.
Figure 2. A histogram of out-degree
We estimated the parameter of a power-law distribution using the poweRlaw library in R, according to the method proposed by Clauset, assuming our data follow a power-law distribution. As a result, out-degrees with values higher than two were estimated to follow the power-law distribution of the following formula (x: out-degree).
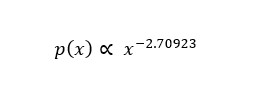
Figure 3. A log-log plot of out-degree
Comparing the distance between the estimated distribution and the 3,000 sampled data from the distribution with the distance between the estimated distribution and our actual data, we could not reject the hypothesis that our data follow a power-law distribution. The P-value was 0.610333, meaning that the distances between 61.03% sets of sampling data and the estimated model were farther than the distance between our actual data and the model.
Finally, we tested the significance of the distance difference between the three alternative distributions and power-law distribution after estimating the parameters of each distribution based on our data: log-normal distribution, exponential distribution, and Poisson distribution. We used Vuong’s likelihood ratio test for this process [17]. The results showed that the power-law distribution had much better explanatory power than the exponential and Poisson distributions. However, the difference in goodness of fit between the power-law and log-normal distribution was indistinguishable. Specific results for each test are shown in Table 1.
Table 1. Vuong's likelihood ratio test.
|
|
p-value |
|
Power-law vs log-normal |
0.6398939 |
|
Power-law vs exponential |
0.006273197 |
|
Power-law vs Poisson |
0.009079904 |
Network structure change depending on time and policy
We analyzed how the network structure changed in accordance with the main policy changes in Korea. Considering Korea’s main quarantine policy, the entire period can be divided into three stages: 1) Early stage (01/20/2020–03/21/2020); 2) social distancing stage (03/22/2020–05/05/2020); and 3) distancing in usual life stage (05/06/2020–7/19/2020).
We measured four indicators for each stage. Two of them are the mean distance and diameter, which were explained in the methods section. The other two are the number of human nodes and the number of links. The first is said to be the number of confirmed patients that make up the infection network at that time, and the latter is the number of infections that occurred at that time. The results are presented in Table 2.
Table 2. Network indicators by period.
|
Period |
Number of human nodes |
Number of links |
Mean distance of the network |
Diameter |
|
Early-stage period |
691 |
250 |
1.447300771 |
6 |
|
Social distancing period |
718 |
133 |
1.204968944 |
3 |
|
Distancing in the usual life period |
1856 |
520 |
1.465359477 |
5 |
It is noteworthy that the mean distance and diameter decreased significantly during the period of social distancing.
The effect of deleting crucial nodes
We analyzed how the overall network structure and key indicators change when important nodes are deleted to anticipate the effects of policies utilizing network information. To this end, we measured several indicators by removing the 32 nodes corresponding to the top 1% and the 164 nodes corresponding to the top 5% based on out-degree. Measured indicators are the four previously utilized indicators. The results are presented in Table 3:
Table 3. Network indicator by removing nodes.
|
Network |
Number of human nodes |
Number of links |
Mean distance of the network |
Diameter |
|
Whole network |
3283 |
1005 |
1.439620081 |
7 |
|
Removing the top 1% |
2786 |
510 |
1.350299401 |
7 |
|
Removing the top 5% |
2383 |
180 |
1.086734694 |
3 |
When the top 1% nodes were deleted, the number of confirmed patients in the network decreased by more than 15%, and by more than 27% when the top 5% were deleted. Figure 4, Figure 5, and Figure 6 present the results of visualizing each network.