Results of NGS analysis
After performing quality-control checks, a total of 52.3 million clean reads were obtained from the raw reads, corresponding to more than 30× coverage of the reference genome. More than 92% of the sequencing data had Phred-like quality scores ≥ 30, indicating that the data were high quality (Table 1). After sequence alignment, nine junction reads on chromosome 03 (Chr03), and four on Chr10, were identified in the Pb29 sequence, indicating that there are two T-DNA insertion sites in the Pb29 genome (Table S1). Based on the physical positions of the junction reads, one insertion site is located at 9,283,937 bp on Chr03, and the other at 10,868,777 bp on Chr10. T-DNA is inserted in the reverse direction on Chr03, and in the forward direction on Chr10. However, further analysis revealed that only unilateral junction reads could be detected at both T-DNA insertion sites; ideally, junction reads should be detected on both sides of each insertion site (Fig. 1).
Table 1 The summary of sequence data from NGS.
|
Clean reads
|
Clean bases (Gb)
|
GC (%)
|
Q20(%)
|
Q30(%)
|
|
52,313,447
|
15.7
|
37.42
|
97.36
|
92.77
|
Confirmation of insertion sites and directions using PCR amplification
To verify the accuracy of the T-DNA insertion sites and directions, we designed 6 primers based on the flanking sequences of the T-DNA insertion sites and the T-DNA sequence (Fig. 2), and amplified the genomic DNA of poplar 741 and Pb29 using different primer combinations (Table 2). The results of PCR amplification revealed that the PCR runs using primer combinations 3, 4, 6, and 7 generated products with a single band for Pb29 in Fig. 3A, whereas no products were amplified for poplar 741 in Fig. 3B. When primer combinations 1, 2, 8, and 9 were used in the PCR, amplified bands were not produced for Pb29 or poplar 741, indicating that T-DNA was indeed inserted into Chr03 in the reverse direction and into Chr10 in the forward direction, thus verifying the NGS results. Meanwhile, the target band was observed after PCR runs using primer combinations 5 and 10 for both Pb29 and poplar 741, indicating that Pb29 is a heterozygous mutant created via T-DNA insertion (Fig. 3).
Table 2 The primer combinations and product size for verifying the insertion sites and directions.
|
No.
|
Primer combination
|
Product size(bp)
|
No.
|
Primer combination
|
Product size(bp)
|
|
1
|
Chr3u-F1 & 131#S5F
|
552
|
6
|
Chr10u-F2 & 131#S5F
|
818
|
|
2
|
Chr3d-R2 & 131#S2F
|
767
|
7
|
Chr10d-R2 & 131#S2F
|
745
|
|
3
|
Chr3u-F1 & 131#S2F
|
671
|
8
|
Chr10u-F2 & 131#S2F
|
937
|
|
4
|
Chr3d-R2 & 131#S5F
|
648
|
9
|
Chr10d-R2 & 131#S5F
|
626
|
|
5
|
Chr3u-F1 & Chr3d-R2
|
684
|
10
|
Chr10u-F2 & Chr10d-R2
|
928
|
Results of Nanopore sequencing analysis
To further verify the NGS results and determine whether chromosomal rearrangement occurred in the Pb29 genome due to T-DNA insertion, we used the third-generation sequencing technology developed by Oxford Nanopore Technologies to resequence the whole genomes of poplar 741 and Pb29. More than 96% of the clean reads of both poplar 741 and Pb29 mapped to the reference genome, corresponding to 40× and 39× coverage of the reference genome, respectively (Table 3). The depth of coverage was evenly distributed across both poplar 741 and Pb29 chromosomes, indicating that the genomic DNA of poplar 741 and Pb29 was sequenced in a random manner (Fig. 4).
The BAM file generated by comparing all junction reads with the P. trichocarpa reference genome was imported into (Integrative Genomics Viewer) IGV software for visual analysis. All junction reads only mapped to Chr03 or Chr10, and there was a gap between reads on both chromosomes. The two gaps, each formed by a T-DNA insertion that disrupted part of the genome sequence, matched the two T-DNA insertion sites in the Pb29 genome exactly. The two T-DNA insertion sites in the Pb29 genome are located at 9,283,905–9,283,937 bp on Chr03 and 10,868,777–10,868,803 bp on Chr10, consistent with the detection results obtained using NGS (Fig. 5).
Compared with the P. trichocarpa reference genome, evidence of many (Structural variation) SV events was seen in the genomes of both poplar 741 and Pb29, most of which were deletions or insertions of chromosome segments (Fig. 6). After removing the regions representing SV events of the same type at the same positions in the poplar 741 and Pb29 genomes, SV events > 1 kb are regarded as chromosomal rearrangements in the Pb29 genome caused by T-DNA insertion. However, we did not detect this type of event, indicating that the insertion of T-DNA did not cause large chromosomal rearrangements in the Pb29 genome.
Table 3 The summary of sequence data from Nanopore sequencing.
|
Sequence Data
|
741
|
Pb29
|
Sequence Data
|
741
|
Pb29
|
|
Clean reads
|
2,351,233
|
2,194,474
|
Clean bases(Gb)
|
20.6
|
19.9
|
|
N50Len
|
10,146
|
10,474
|
N90Len
|
6,147
|
6,414
|
|
MeanLen
|
8,778
|
9,072
|
unmapped
|
88,268
|
83,507
|
|
mapped
|
2,262,965
|
2,110,967
|
Mapped ratio(%)
|
96.25
|
96.19
|
|
Average depth
|
40
|
39
|
Coverage_ratio_1X(%)
|
84.99
|
84.8
|
|
Coverage_ratio_5X(%)
|
75.23
|
74.92
|
Coverage_ratio_10X(%)
|
69.49
|
69.12
|
T-DNA and flanking sequence analysis
Because Nanopore sequencing can be used to obtain longer reads, some junction reads contained complete T-DNA sequences. The complete T-DNA sequences at the two insertion sites were extracted and compared with the vector sequence. The results showed that the left and right border sequences of the T-DNA inserted on Chr03 were missing 26 and 3 bp, respectively, whereas the left and right border sequences of the T-DNA inserted on Chr10 were missing 35 and 34 bp, respectively (Fig. 7). It is worth noting that the 35S-API-Nos expression component was not detected in the T-DNA sequences at either insertion site; furthermore, both T-DNA sequences are exactly the same, indicating that the expression component of the API gene was not lost during the transformation process. Rather, it was not present in the expression vector in Agrobacterium before transformation (Fig. 8).
We compared isolated flanking sequences with the P. trichocarpa reference genome and found that fragments had been deleted from the flanking sequences at both insertion sites, as T-DNA insertion damaged the genome sequence at those sites (box with red outline in Fig. 9). The genome sequence at the T-DNA insertion sites on Chr03 and Chr10 was missing 33 and 27 bp, respectively, consistent with the results of the alignment analysis (Fig. 5). A short fragment (24 bp in length) was found between the T-DNA insertion site and the right flanking sequence on Chr03 in the Pb29 genome; this fragment could not be mapped to the P. trichocarpa reference genome. We analyzed the clean reads from poplar 741 found that reads mapped to the same positions essentially had the same sequences as the corresponding sections of the P. trichocarpa genome (Fig. 10), indicating that the 24-bp fragment did not arise from the difference between genomes but was instead caused by the insertion of an unknown fragment during the T-DNA integration process.
Analysis of the expression levels of genes located near the insertion sites
The genes within 20 kb upstream and downstream of the two T-DNA insertion sites were detected based on the genome annotation file of P. trichocarpa. The results showed that T-DNA was inserted 9,466 bp downstream of the LOC112326972 gene and 8,137 bp upstream of the LOC7475699 gene on Chr03, and 15,621 bp downstream of the LOC7498060 gene and 1,543 and 11,914 bp upstream of the LOC7498061 and LOC7498062 genes, respectively, on Chr10 (Table 4). (Fragments Per Kilobase Million) FPKM values associated with the transcriptome data were used to compare the expression levels of the five neighboring genes. The results showed that except for the LOC7498061 gene, the expression levels of the other four genes in Pb29 leaves did not change significantly, indicating that the insertion of T-DNA did not significantly affect the expression levels of these four genes. The LOC7498061 gene is located closest to the T-DNA insertion site; its expression level was significantly upregulated in Pb29 leaves, indicating that the insertion of T-DNA in Pb29 affects gene expression within a certain range (Fig. 11).
Table 4 The genes located near the insertion sites.
|
Insertion location
|
Neighboring gene(< 20 kb)
|
Genomic location
|
|
Chr03:9283905-9283937
|
Upstream
|
LOC112326972
|
Chr03:9261716:9274439
|
|
Downstream
|
LOC7475699
|
Chr03:9292074:9294391
|
|
Chr10:10868777-10868803
|
Upstream
|
LOC7498060
|
Chr10:10848741:10853156
|
|
Downstream
|
LOC7498061
|
Chr10:10870346:10873516
|
|
LOC7498062
|
Chr10:10880717:10883716
|
Analysis of the TAFs gene family
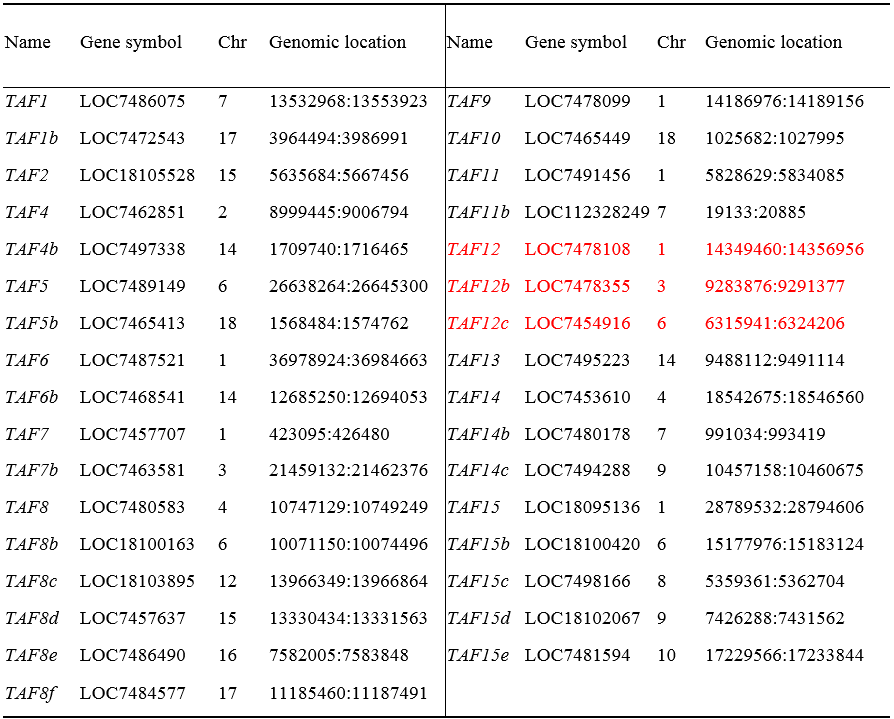
According to the results of whole-genome resequencing analysis, the T-DNA insertion site on Chr03 (9,283,895–9,283,937 bp) is located within the first exon of the LOC7478355 gene (9,283,876–9,291,377 bp). Therefore, the insertion of T-DNA disrupted the structure of the LOC7478355 gene. According to the National Center for Biotechnology Information (NCBI) analysis, the LOC7478355 gene, which belongs to the TAFs gene family, encodes a TAF12 protein, which is one of the core subunits constituting the basic transcription factor TFIID. To understand the impact that this disruption of the gene structure has on the function of this gene, we first analyzed the TAFs gene family to clarify the number of genes encoding TAF12 protein in the genome.
We identified 33 TAFs genes in the genome of P. trichocarpa through bioinformatics analysis. The 33 PtTAFs genes were renamed according to their chromosomal positions and the phylogenetic tree constructed with PtTAFs and AtTAFs proteins (Table 5; Fig. 12A). Within the TAFs gene family, there are three genes encoding TAF12 protein PtTAF12, PtTAF12b, and PtTAF12c. Through synteny analysis of the PtTAFs gene family, we identified five segmental duplication events involving 10 PtTAF genes that encode TAF7, TAF8, and TAF15 proteins. No duplicated segments containing genes encoding TAF12 protein were identified, indicating that PtTAF12, PtTAF12b, and PtTAF12c were not formed from segmental duplication occurring among the three genes (Fig. 12B). The RNA-seq results showed that the expression levels of the three genes in Pb29 leaves were slightly higher than those in poplar 741, but none of the differences were significant, indicating that the transcriptional abundance of the genes encoding TAF12 protein did not change significantly (Fig. 13).
Table 5 The physical characteristics of TAFs gene family in Populus trichocarpa.