Participants and Inclusion/Exclusion Criteria
Subjects were recruited through posted flyers and emails and from inpatient rehabilitation, stroke recovery, and neurosurgery units at the Strong Memorial Hospital in Rochester, NY. Inclusion Criteria (for subjects with history of stroke) included having moderate UE weakness (Medical Research Council (MRC) strength scale score: 3-4) due to recent stroke. Patients with both ischemic and hemorrhagic strokes were recruited for this study. Exclusion criteria (for both healthy controls and subjects with recent stroke) included chronic UE injury or pain. Patients with history of recent stroke were also excluded if they had severe UE weakness (MRC<2), aphasia/cognitive impairment such that the patient was not independently making choices about their healthcare decisions, or if their health care provider determined they were unable to safely participate in rehabilitation therapies.
Sensor Placement and Data Acquisition
Three MC10 BioStampRC® wearable sensors were placed on the subject’s affected arms to record a combination of either triaxial accelerometry and electromyography or triaxial accelerometry and gyroscope data during three prompted UE exercises and resting periods. In this manuscript, only results using accelerometry and gyroscopy data are discussed.
One sensor each was placed on the upper arm (volar surface of the brachium), forearm (medial volar surface), and hand (dorsal surface) (Fig. 1). The forearm sensor was originally placed on the medial forearm over the bulk of the wrist flexor muscle group (Fig. 1A). However, when put into practice with inpatient subjects, this sensor placement was periodically inconvenient due to the presence of peripheral intravenous lines or large, painful bruises that formed after the removal of such lines from the antecubital region. Thus we also collected data from a more lateral position on forearm, over the wrist extensor muscle group (Fig. 1B), to ensure that the system could still be applied for a patient if the antecubital fossa region were unavailable for sensor placement (for example, when a line is placed in the paretic arm in the setting of emergency medical treatment or because both arms are paretic).
Exercise Protocol
The subjects received verbal instructions and visual demonstrations on how to perform three exercises (Fig. 1 C, D, and E):
- Exercise 1: flexion/extension of the elbow (Fig. 1C)
- Exercise 2: supination/pronation of the forearm (Fig. 1D)
- Exercise 3: extension/flexion of the wrist (Fig. 1E)
Patients were asked to perform 20 repetitions of each exercise per session, with a total of three exercise sessions separated by at least 20 minutes. To ensure accurate movements, study personnel supervised the exercise activities during the exercise session and provided additional demonstrations of the correct exercise movements as needed. They also tagged the start and stop of each exercise type on the MC10 application, so that the “true” activity was labeled at the correct time on the data recording. Sensors were kept on subjects’ arms for one to three hours in order to record data corresponding to resting and non-exercise movement patterns and confirm that the algorithm could effectively discriminate and ignore non-exercise arm movements. Patients were allowed to perform any paretic arm movement they wished during the resting periods but were asked to avoid movements that replicated any of the three study exercises. Recording sessions were timed such that the subject did not have occupational or physical therapy rehabilitation sessions during the data recording period.
Data Processing
Data were recorded simultaneously from the three sensors. Since data sets from different sensors are not perfectly synchronized and are sampled at different sampling frequencies, post-processing was required to ensure each raw data set could be used to train the machine learning-based classification algorithm. MATLAB and Delimit software programs were used for all data processing. The first step involved linear interpolation of the data to match sensor sampling frequencies, which may differ depending on the recording mode. This was done in MATLAB using the interp1 function. In the second step, data was synchronized by the first Unix timestamp value that was associated with each period in the exercise session. The data was then labeled according to Unix timestamp intervals, which were created by a start/stop timer included in the MC10 application. Data sets were filtered using a bandpass filter (0.1 to 1.5 Hz) to remove high-frequency noise and wandering baselines due to gravity signatures, (changes in baselines due to the force of gravity acting on the sensors differently, depending on their orientation) then z-score normalized in MATLAB using the normalize function.
Classification, Data Extraction, and Repetition Enumeration Using Peak Finding
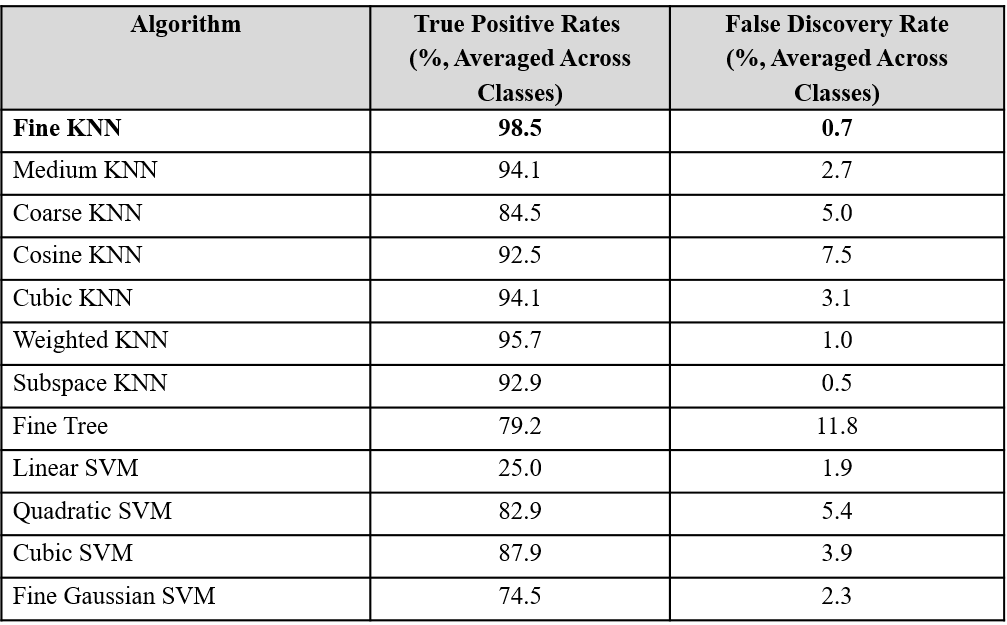
Once all data sets were synchronized, labelled, filtered, and normalized, the Fine k nearest neighbor (k-NN) classification algorithm provided by MATLAB was used to classify the data using a leave-one-out cross validation method. This was accomplished by training the algorithm on all datasets except for one, and then testing the algorithm on the “left-out” dataset. This test produced a set of algorithm-predicted labels for each row of the “left out” dataset. The Fine k-NN algorithm was utilized over other possible classification algorithms because when applied to the data in MATLAB’s classification learner add-on, the Fine k-NN algorithm achieved the highest reported classification accuracy (Table 1).
Table 1: Comparison of classification accuracy on all subject data using various classification algorithms. Multiple types of MATLAB classification algorithms were trained and tested using a leave-one-out cross validation method, and the mean classification accuracy rate for each classification algorithm were compared. The fine k nearest neighbor algorithm provided highest true positive rates and lowest false positive rate, and was therefore chosen for subsequent analyses.
To calculate the number of repetitions a subject performed of each exercise, data corresponding to Exercise 1, Exercise 2, and Exercise 3 were separately extracted from each dataset according to the column of algorithm-predicted discrete labels, and new tables of each activity were created for each subject. The MATLAB findpeaks function was then applied to different variable signals in each of these extracted activity tables to count the number of repetitions for each activity. For the Exercise 1 (elbow flexion/extension) tables, the x-axis accelerometer data from the forearm sensor was used, because clear peaks representing Exercise 1 repetitions are found in this data. For the same reason, the y-axis and x-axis gyroscope signal from the dorsal hand sensor were used to count repetitions of Exercise 2 and 3, respectively. An example of this process, as well as the previously mentioned data pre-processing methods, are shown in Figure 2.
Peak Counting Accuracy Calculation
To determine peak counting accuracy, the system’s estimate of the number of repetitions performed by the patient for each exercise, Nautomatic, was compared to the repetition number as reported by the study personnel, Nmanual. To calculate this accuracy, the percent error was calculated for each count and subtracted from 100%, according to the following formula:

